Commercial industrial engineering is the savior of high-throughput genomics
We ‘completed’ the human genome in 2003, thanks mostly to commercial industrial engineering.
Genome sequencing didn’t really enter the household vernacular until the announcement of the Human Genome Project in the early 90’s, and that was only possible because of major advancements in sequencing technology.
For the better part of a decade, Sanger sequencing was performed using radiolabeled nucleotides and chain terminators, one for each base, requiring a lane for each A, C, T, and G.
In 1987, George Trainor and a team at DuPont got the bright idea, literally, to use fluorescently labeled chain-terminators, enabling the multiplexing of the Sanger sequencing reaction into a single tube.
This might sound like a simple and obvious feat, but it wasn’t!
The advancement here isn’t just “add fluorescently tagged bases and BOOM! Here’s the data.”
They had to find a compatible polymerase, synthesize the dyes, tag the nucleotides, and engineer an optical system for data acquisition.
The system itself is an engineering wonder: there’s a laser, a beam expander, a suite of scanning optics, two detectors with their own filter stacks, all aligned to excite and detect the dyes in the sequencing gel bands as they pass across a laser beam.
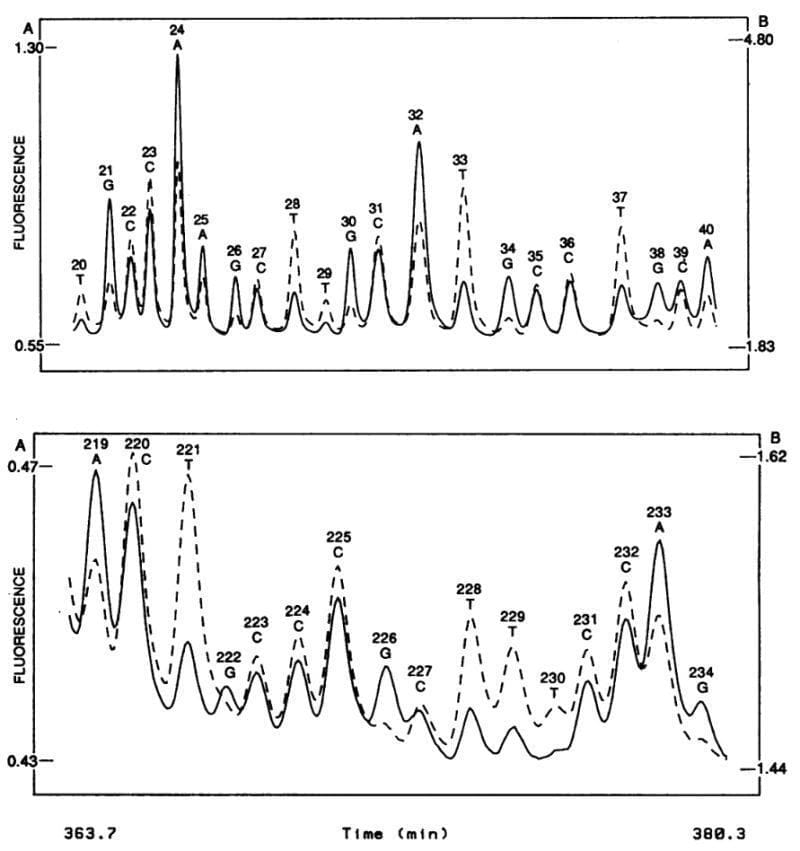
The figure above shows what this groundbreaking system was able to accomplish - the sequence of two 20 base pair stretches of M13mp18, a lac phage vector.
The solid line is the fluorescence intensity observed by the A, G detector and the dashed line the intensity observed by the C, T detector.
These plots should look suspiciously familiar to anyone that’s done fluorescence based Sanger sequencing and it probably isn’t a surprise that, through a licensing agreement, the principles of the technology in this paper made their way into the first four color commercial sequencer in 1988, the ABI 370A.
Despite this process remaining gel based: multiplexing the reaction, eliminating the radioactive nucleotides, ditching the film, and automating the acquisition of the sequencing data were all a huge triumph.
But because it still required the pouring of a gel, it gave grad students and postdocs something to complain about for at least another decade; unfortunately for them, the capillary electrophoresis based ABI Prism line wasn't introduced until 1996.
Read the full issue of Omic.ly Premium 18