I sometimes give Illumina a hard time, but they've commercialized some killer products
Illumina is considered by many to have single handedly transformed the field of genomics. They did it by building on the foundation Solexa established in 1997.
A lot of great DNA stories start in an English pub and this one isn't an exception.
Shankar Balasubramanian and David Klenerman formed Solexa after deep discussions at the Panton Arms Pub sparked a new idea for how to revolutionize DNA sequencing.
Their concept centered on the visualization of single DNA molecules as they incorporated nucleotides on a solid surface.
This sounds deceptively simple.
But, key components of this technology included the development of a reversible sequencing chemistry, a method for imaging single molecules, and a highly parallelized array with the goal of sequencing over 1 billion bases.
Sequencing single molecules proved to be a huge challenge but Solexa was able to pivot their approach by acquiring a clustering chemistry from a struggling Swiss sequencing firm named Manteia.
Their chemistry was conceptualized by Pascal Mayer and Laurent Farinelli and it created spots of ~1,000 identical molecules.
This was significant because it amplified the signal that Solexa was able to detect during each sequencing cycle.
With this foundation, they maintained a competitive edge by continuously improving on their quality.
As a result, Illumina acquired them in 2006 for $650m.
Incredibly, 2 years later, they published a full human genome sequence.
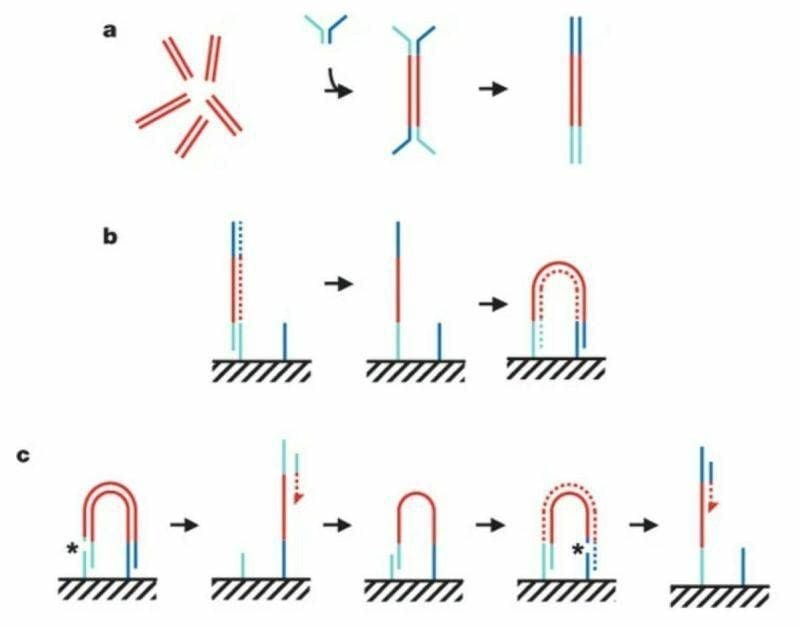
The figure above summarizes the sequencing chemistry described in that seminal paper:
Illumina-Solexa libraries are made by breaking DNA into short ~300bp fragments, ligating on adapters, and then amplifying with PCR as seen in (a).
The adapter ligation step is crucial for 2 reasons 1) it adds known sequence to an unknown fragment to amplify it with PCR but 2) that known sequence can be used to capture those molecules as seen in (b).
Capturing these molecules on a slide is critical for the process of clustering.
In (b) you can see the turquoise adapter binding to a complementary sequence on a slide, it’s copied through an extension reaction, and then 'bridges' over to another complementary sequence where the process is repeated.
This is done multiple times to create a lawn of clonally amplified clusters.
Sequencing occurs through the addition of a sequencing primer, depicted here as a turquoise bar with a red dashed arrow.
This is followed by another round of clustering and bridging to sequence the opposite end of the DNA, depicted now as the blue bar with the red dashed arrow.
In the 15 years since the publication of this paper, millions of people have been sequenced using this technology.
It's hard to imagine that the genetic revolution would have been possible without these groundbreaking innovations.