The human genome sequence is finally (almost) totally complete. The pesky Y chromosome was the last hold-out!
The human Y-chromosome has finally been fully sequenced! No, seriously this time, the genome is actually finished now, maybe.
You might be asking yourself why we keep hearing about the completion of the human genome.
It's like the scientific gift that keeps giving and we're probably going to continue reading these stories because nothing in science is ever really finished.
The more we dig, the more we find, and that presents new questions and generates new hypotheses!
So why'd it take so long to finally sequence the Y chromosome?
Well, over half of it is composed of highly repetitive sequence that couldn't be resolved with short-read sequencing or Sanger sequencing.
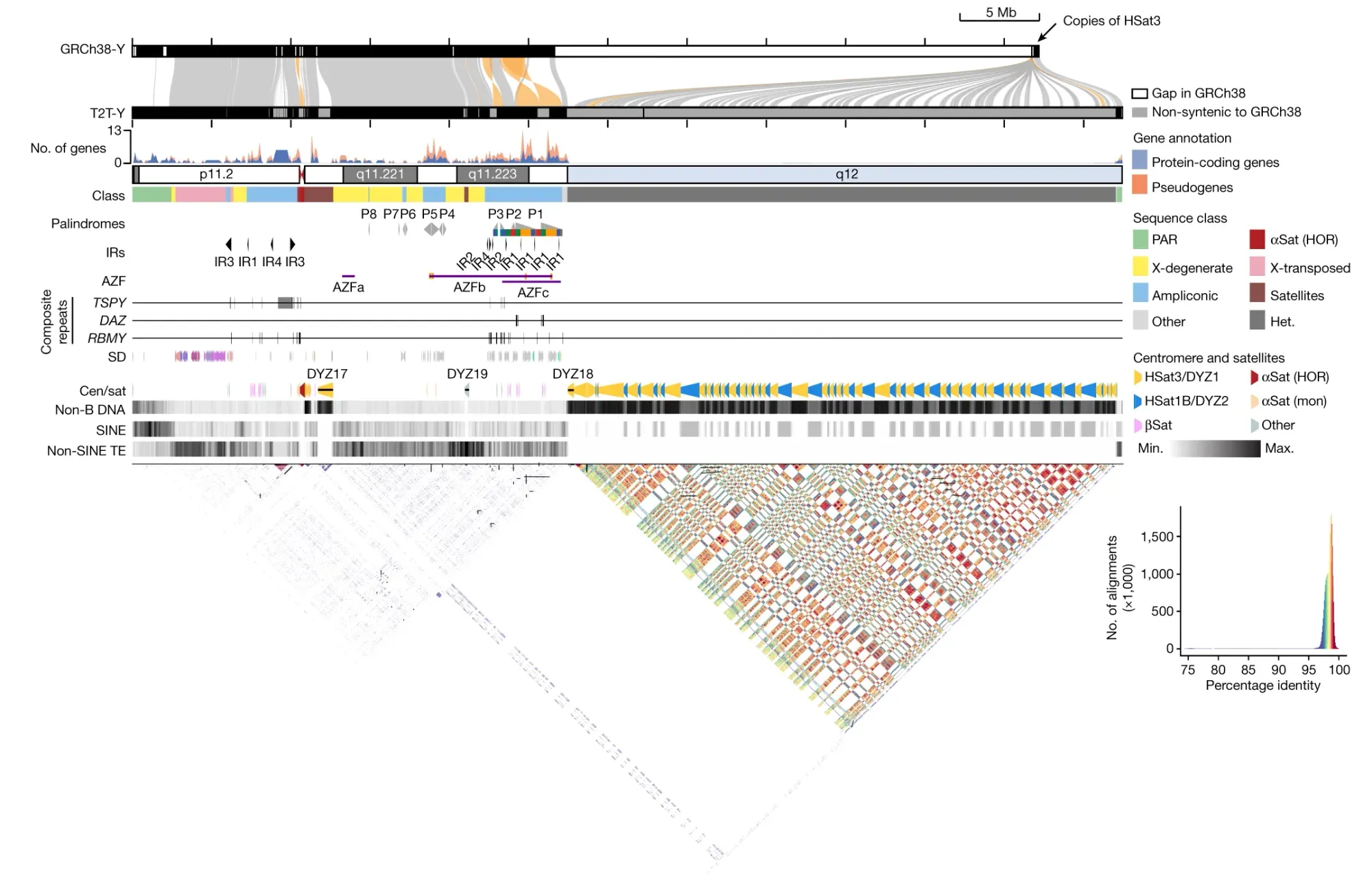
Gaps have persisted due to the complex architecture of Y, including palindromes and repetitive sequences which make assembly of these sequences challenging.
Fortunately, in the decades since the genome was 'finished' the first time, we've developed new long-read methods that can span these complicated regions and they were used in combination with short-read polishing to generate the latest reference.
Short of a couple small regions that still need some attention, it's finally done!
This is important because the human Y chromosome and its SRY locus is what determines sex in mammals (whether you are biologically male or female).
This is of particular interest in the case of evolution and sex determination because a number of non-mammalian species have lost their Y chromosome and this latest work could help us to understand how mammals could suffer a similar fate.
To that end, the Telomere-to-Telomere (T2T) consortium successfully assembled the complete Y chromosome of HG002, T2T-Y.
The final assembly was 62,460,029 bases in length and shows approximately 30 Mb of previously uncharacterized sequence, predominantly from the heterochromatic region of the q-arm.
The researchers further analyzed the gene content, repeat sequences, centromeric regions, and other features of T2T-Y.
They identified, "an additional 110 genes, among which 41 are predicted to be protein coding. The majority of these protein-coding genes (38 of 41) are additional copies of TSPY."
The paper went on to confirm that the number of copies of TSPY and a handful of other repetitive genes can vary greatly between individuals (a second paper in Nature dives into this in-depth).
The researchers also characterized new complex structural features on the Y chromosome (see the figure below), and improved variant calling for XY individuals using T2T-Y as a reference, showing enhanced accuracy in variant detection and reducing the number of false-positive variant calls.
The completion of T2T-Y and the availability of complete genome references from diverse populations are the cornerstones of advancing our understanding of human genetics.
Read the full issue of Omic.ly Premium 13