Frederick Sanger sequenced the first DNA genome, he didn't use 'Sanger Sequencing' to do it
Frederick Sanger invented a famous DNA sequencing method. It's not the one pictured below (which he also invented).
Sanger is one of the few scientists to be awarded more than one Nobel Prize.
His first was for determining the amino acid sequence of Insulin!
His second was shared with Walter Gilbert and Paul Berg for developing nucleic acid sequencing methods.
To this day, 'Sanger' sequencing is a gold standard method in genetics.
And despite the advent of high throughput sequencing technology, millions of Sanger reactions are still performed every year!
But Sanger has another claim to fame.
He sequenced the first DNA genome which was the bacteriophage PhiX174.
This may sound familiar.
Because PhiX is used widely as a sequencing control!
This is partly to memorialize that it was the first, but mostly because it has a similar number of AT and GC bases.
But, Sanger didn't use his eponymous sequencing method to sequence it!
This came as a bit of a shock to me.
And my discovery of this fact went something like this:
“What the (expletive) is the ‘plus and minus’ method!?”
And so, I'm not going to cover Sanger's sequencing of bacteriophage PhiX174.
Instead, we'll review the method he used to do it!
So, what exactly is the 'plus and minus' method?
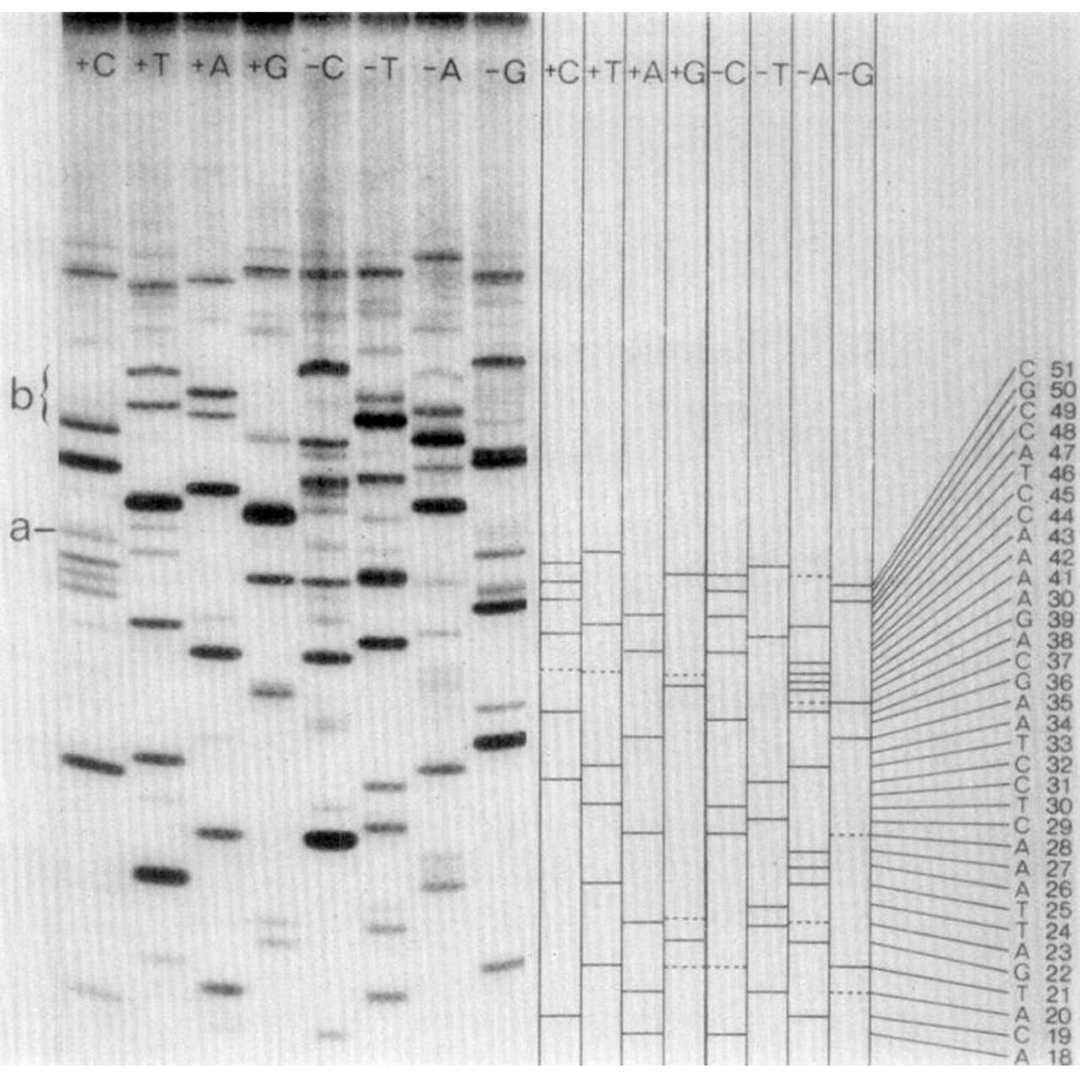
It can be seen above.
But don't be fooled!
While it looks like regular old gel based Sanger sequencing, there are actually twice as many lanes!
And they're labeled ‘+C+T+A+G’ and ‘-C-T-A-G.’
Are you starting to see where the 'plus and minus' name came from?
The minus lanes are made by priming the thing you want to sequence with a restriction fragment, adding all the bases and a polymerase, and then stopping the reaction.
Those random products are then purified and added to a new reaction that contains only 3 of the 4 bases.
Whatever additional extension occurs stops at the position of the missing base.
So, the band in the gel is -1 from where the -base should be!
The plus lanes are made using a similar process:
Random fragments are generated and then the purified fragments are added to a new reaction containing only 1 of the nucleotides along with T4 polymerase.
T4 polymerase is special because it will 3’ degrade DNA, but interestingly, it will stop degrading DNA at “residues corresponding to the one [nucleotide] that is present.”
Translated: if you only add adenine nucleotides, it creates fragments that all end in adenine.
So, when you run these 8 reactions (+C+T+A+G -C-T-A-G) out on a gel, you can use the plus and minus signals from each base to determine the sequence of the fragment that was present!
Two years later, chain terminating dideoxynucleotides were added to this scheme, allowing 'Sanger' sequencing to be done in 4 lanes instead of 8 and with fewer steps and artifacts.

