The 'Histone Code' fundamentally changed how we think about gene expression
Histones were once thought to just be dumb structural proteins that held chromatin together; now we know that the ‘histone code’ is a major regulator of gene expression.
The first studies on gene expression were done in bacteria.
Here, gene expression is very straightforward:
Proteins bind to DNA, turn it into RNA and then that RNA gets read to create other proteins.
Things get much trickier in complex organisms.
Their larger genomes are packaged into a super-structure called chromatin.
And, in 1974, Roger Kornberg proposed that chromatin was made up of a DNA-protein complex that he named the ‘nucleosome.’
DNA is wrapped around histone proteins to create this highly ordered structure but this also presents an interesting problem to solve.
If DNA is all coiled up, how does the gene expression machinery get in there to keep cells alive?
Fortunately, David Allis suspected that histones were critical players in gene regulation.
He remembered earlier work from Vincent Allfrey showing that histones could be methylated or acetylated and he was determined to figure out if these modifications performed a function.
His chosen model for this was the single-celled protozoan Tetrahymena.
This might seem an odd choice, but Tetrahymena are full of acetylated histones and have high levels of gene expression which made them perfect for the task!
Allis and his team figured an enzyme was responsible for all of this acetylation and isolated the first histone acetyltransferase (HAT) from them in 1996.
They found that this enzyme, p55, was similar to Gcn5p, a protein known to regulate gene expression in yeast.
This discovery, along with additional work from Michael Grunstein, connected HATs to gene activation.
But adding an activation mark to histones only solves half of the gene expression problem.
How do you turn genes off?
Fortunately, the first histone deacetylase (removes acetyl groups) was discovered soon after by Stuart Schreiber and this explained how acetylation levels were maintained in cells.
Following these two discoveries, additional histone modifying enzymes were found that added phosphorylation and methylation to the mix.
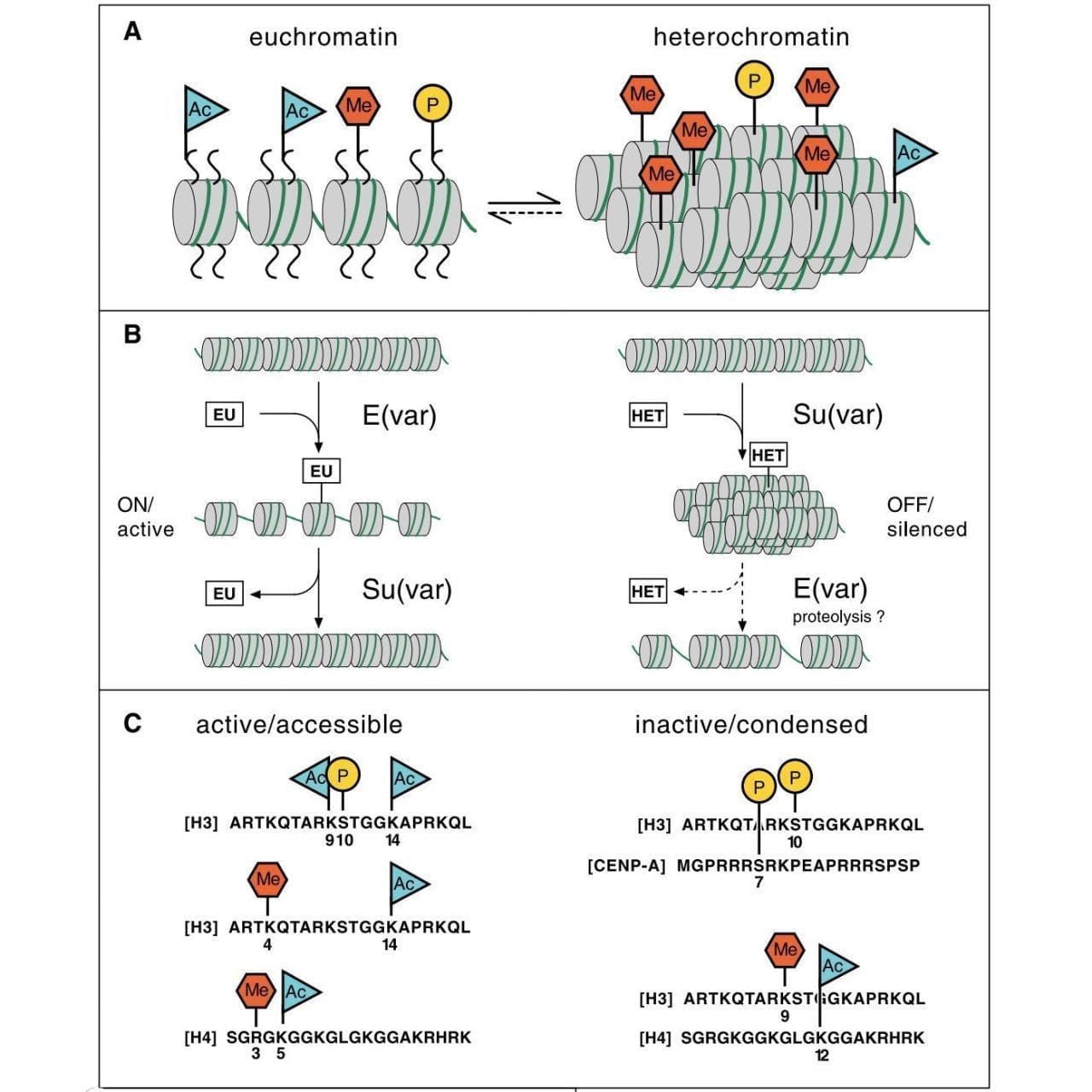
The figure above pulls all of this together into what is known as the histone code, or, how histone modifications are added, read, and erased to control what regions of DNA are accessible for gene expression.
A) displays the histone marks associated with open (euchromatin) and closed (heterochromatin), B) shows a model of how suppressors and enhancers of variegation work to open and close chromatin, and C) shows which amino acids on each histone are marked in open and closed chromatin.
But, this seminal article is important because it is one of the first to link histone modifications to epigenetics and this idea fundamentally changed how we viewed gene regulation at the chromatin level, forever.

