A wormy discovery becomes a knockout laboratory technique
One of the best ways to figure out what a gene does is to get rid of it and see what happens.
Prior to 1998, this was time consuming and tedious.
That all changed with the discovery of RNA interference.
Gene expression is the process by which the genetic code is converted into something that performs a function within the cell.
Most of the time, this means the conversion of DNA into messenger RNA and then messenger RNA (mRNA) into protein.
It had been known for some time that gene expression could be modified through the introduction of 'anti-sense' or complementary RNA. It was thought that this RNA would bind to the mRNA and prevent it from being read to create protein.
While useful in some biological studies, the effect of anti-sense RNA treatment was modest and the mechanism of how it worked was poorly understood.
So, in 1998, Andy Fire and Craig Mello embarked on a journey to see if they could make this gene expression modification tool more efficient, hypothesizing that the structure of the anti-sense RNA might play a key role.
What they discovered surprised them.
Being good scientists, they introduced anti-sense RNA, sense RNA and both anti-sense and sense together as a double-stranded RNA (dsRNA) complex to determine what effect these combinations would have on gene expression.
The sense and anti-sense treatments had modest effects on their study subject, the nematode worm, C. elegans, but the inclusion of the dsRNA combination had a striking phenotypic effect, mimicking what would have been expected if the genes they were targeting were totally knocked out.
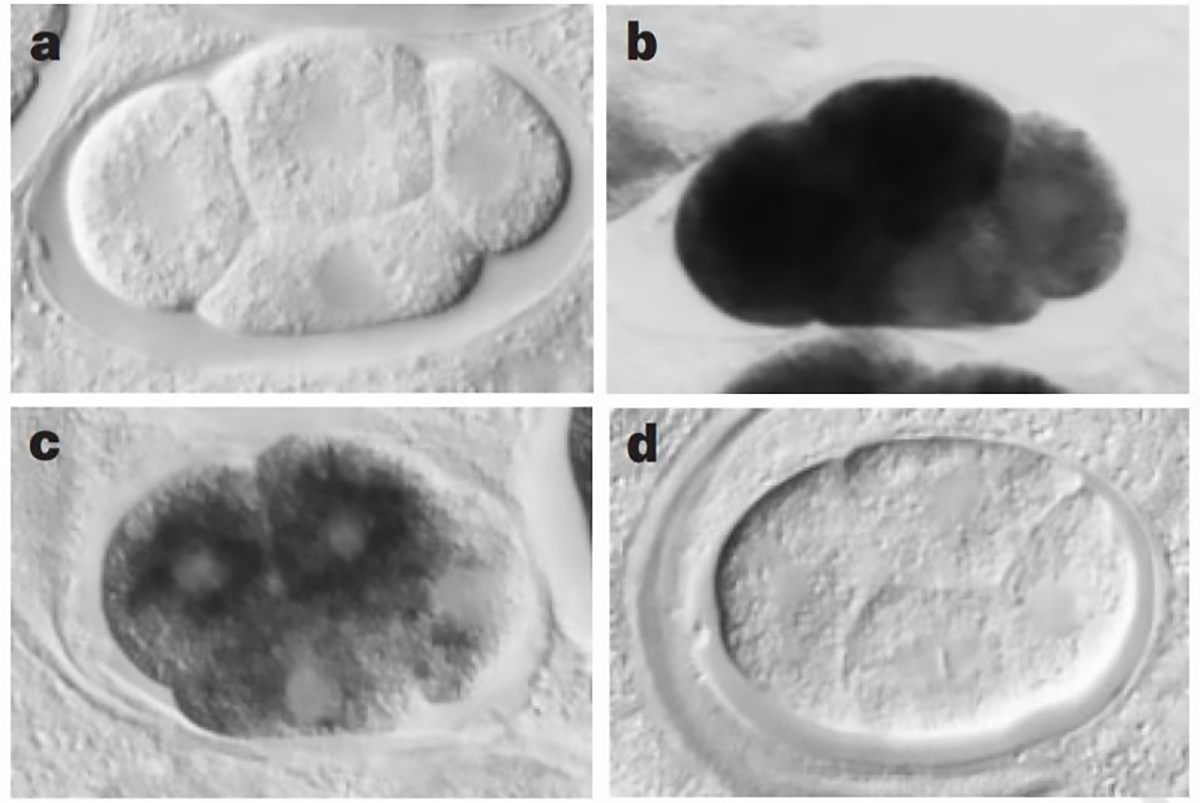
The figure above shows a series of images of a C. elegans embryo. Panel A is the negative control; Panel B shows the staining of an abundant mRNA, mex-3; Panel C shows what happens to mex-3 RNA in the presence of anti-sense RNA; and D shows the complete obliteration of mex-3 RNA after the injection of dsRNA.
Further work after this 1998 paper resulted in the discovery that RNA interference is actually a biological process that cells use to regulate the expression of genes, allowing scientists to identify and name a bunch of new proteins like Drosha, Dicer, Argonaut and the RNA-Induced Silencing Complex (RISC).
We have since co-opted this process for our own uses as a scientific tool and also as a therapeutic for treating human diseases.
Unsurprisingly, given its utility and continued impact, Andy Fire and Craig Mello received the Nobel Prize in Physiology or Medicine in 2006 for their discovery of RNA interference.
