A USB drive? Nah, an epigenetic drive!
DNA data storage is coming for all of your bits (but now with an epigenetic spin)!
Deoxyribonucleic acid, which we lovingly refer to as DNA, is the ultimate biological data storage material.
It stores most of the information required for us to function within its sequences of adenine, thymine, guanine, and cytosine!
And because DNA is so small, it has been the envy of computer scientists who see it as a potential solution to our impending data storage dilemma.
We’re currently creating over 400 terabytes (TB) of data a day.
A typical 15 TB data storage tape weighs 200 grams which comes out to roughly 0.075 TB per gram.
DNA can store 215 Petabytes (PB) per gram.
A PB is equal to 1000 TB.
So, DNA can store *checks math* 13,333 times more data than a typical tape drive!
You can see why we might be interested in using DNA to help satiate our data storage needs.
But, up until now, all discussions of DNA based data storage have been around methodically synthesizing long stretches of DNA to encode information.
This is very expensive, slow and hard to scale.
Fortunately, there’s a relatively unexplored alternative DNA based storage method that our cells use every day that doesn’t require the complicated and methodical synthesis of DNA.
You might remember that the DNA bases themselves don’t provide all of the data that’s required for our cells to function.
There’s a lot of information encoded in the non-sequence based parts of our DNA and epigenetic modifications to DNA bases could potentially be used for data storage!
This would allow us to encode information into bits of DNA using things like DNA methyltransferases instead of having to bulk synthesize new sequences.
And because sequencers from PacBio and Oxford Nanopore can natively read epigenetic modifications now, we have high quality epigenetic data retrievers at our disposal!
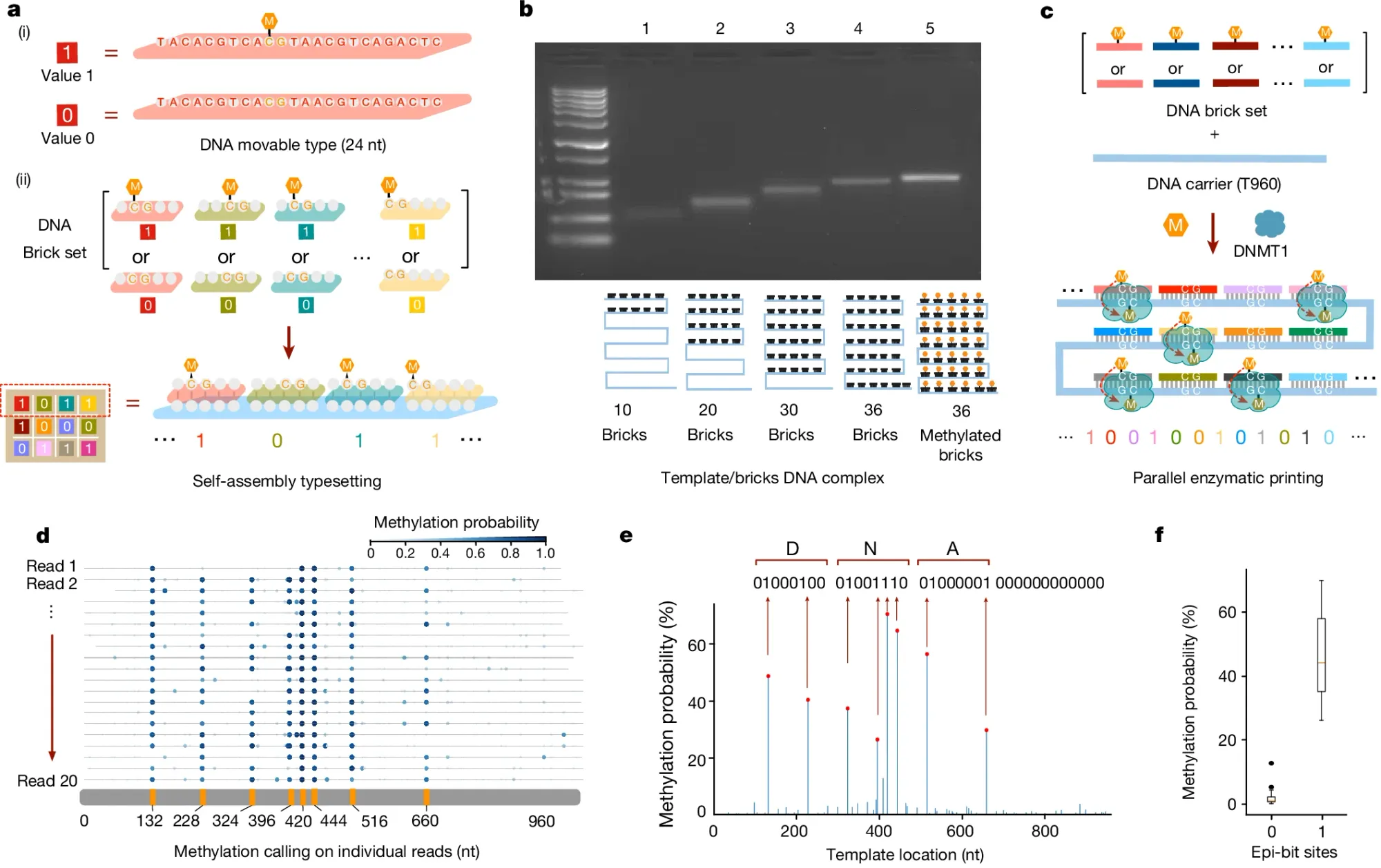
In the figure above, the researchers behind this week’s paper leveraged an additional emerging technology to perform this task: DNA self-assembly.
a) They used 24 base pair DNA ‘bricks’ to represent binary 1 or 0 codes and the self-assembly (hybridization) of these methylated (1) or unmethylated (0) bricks to a complementary template is how they began the process of encoding the data into the template
b) shows they can detect size and methylation differences of template hybridized bricks on a native gel
c) explains how a DNA methyltransferase (DMNT1) is used to copy the methylation signal from the bricks to the template
d) highlights how methylation aware sequencing of the methylated template is used to read back the encoded data
e) is a graph of the final consensus calls for the methylation status of each brick (‘DNA’ was spelled in binary code)
f) displays the error of the methylation calls (no methylation has much lower error than methylation calls)
The researchers went on to show they could store complicated information within this ‘epi-bit’ system, storing an image of a tiger, a panda, and encoding large blocks of text.
However, this system wasn’t perfect and in the worst case had an error rate at data retrieval of nearly 10%.
Errors can be introduced in multiple places including during encoding, storage (methylation signals can be lost), and errors during sequencing.
These should all improve as the technology is developed further.
But, as a proof-of-concept, it’s quite exciting to see how we might finally be able to use DNA to efficiently and cost effectively store more than just biological data!
